Part Number: DLPNIRSCANEVM
Other Parts Discussed in Thread: DLPC350, ADS1255,
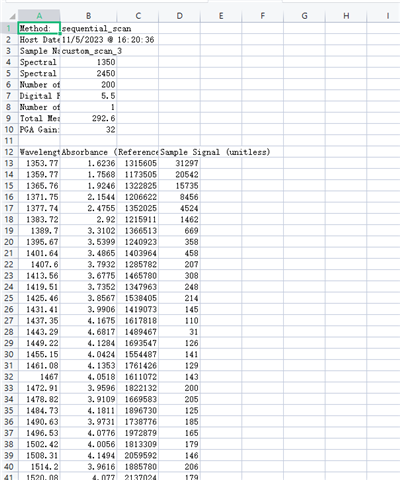
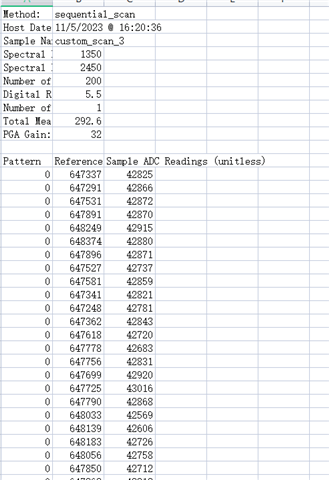
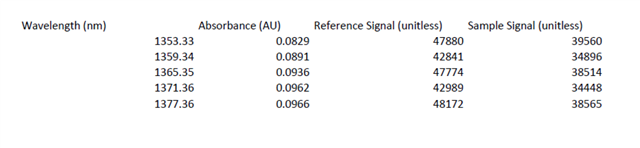
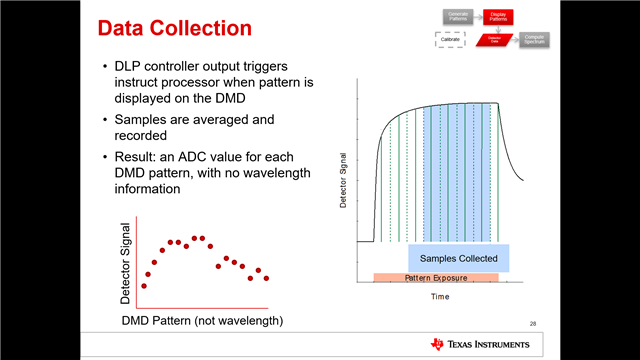
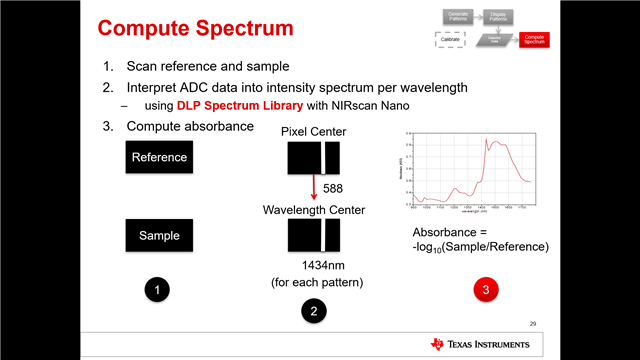
使用DLP的custom_scan功能进行测量时,得到的Spectrum Data和Raw ADC Data两组数据之间的联系是如何的呢?Raw ADC Data经过怎样的处理得到的Spectrum Data?我查阅使用手册,手册中说Spectrum Data是由平均之后的数值,原文是“Whereas the Download Spectrum button provides averaged reference and sample data values”,但我用Raw ADC Data进行平均得到的结果与Spectrum Data相差甚远。